library(tidyverse)
library(AMCP)
library(dfmtbx)Two-way ANOVA
Building off of the one-way ANOVA, a two-way ANOVA is an extension for situations where an additional between-subjects factor is of interest.
Data set description
The example data set comes from table 5 of chapter 7 from the AMCP package. In this data set, a hypothetical experiment tests the effect of presence or absence of biofeedback (first between-subjects factor) in combination with three different drugs (second between-subjects factor) on measures of blood pressure (outcome/dependent variable). The Feedback group is coded as a 1 or a 2 where 1 indicates the participants received biofeedback and 2 indicates participants did not. Drug is coded as 1, 2, or 3, and specifies one of three hypothetical drugs that purportedly reduce blood pressure. Finally, Scores refer to the dependent variable and measure blood pressure where lower values are better. This leaves us with a 2x3 between-subjects design. We will also assume that the assumptions of independence, equality of variance, and normality are met for the sample data set.
Prepare the data
For this approach we will want to convert the Feedback and Drug variables into factors to prevent R from treating them as numerical variables. We will also assume that the assumptions of independence, equality of variance, and normality are met for the sample data set.
# Load data
data("chapter_7_table_5")
data <- chapter_7_table_5
# Convert Group to factor
data <-
data %>%
mutate(across(Feedback:Drug, ~ factor(.x)))
# Display the first 6 rows of the data
data %>%
head() %>%
format_gt_tbl()| Score | Feedback | Drug |
|---|---|---|
| 170 | 1 | 1 |
| 175 | 1 | 1 |
| 165 | 1 | 1 |
| 180 | 1 | 1 |
| 160 | 1 | 1 |
| 186 | 1 | 2 |
Generate summary statistics and plots
By Drug
# Summary statistics
data %>%
group_by(Drug) %>%
summarise(
n = n(),
min = min(Score),
max = max(Score),
median = median(Score),
mean = mean(Score),
sd = sd(Score)
) %>%
format_gt_tbl()| Drug | n | min | max | median | mean | sd |
|---|---|---|---|---|---|---|
| 1 | 10 | 160 | 197 | 175.50 | 178 | 12.29 |
| 2 | 10 | 186 | 219 | 200.00 | 202 | 11.84 |
| 3 | 10 | 170 | 228 | 200.50 | 199 | 18.18 |
data %>%
ggplot(aes(x = Drug, y = Score)) +
geom_boxplot() +
theme_minimal()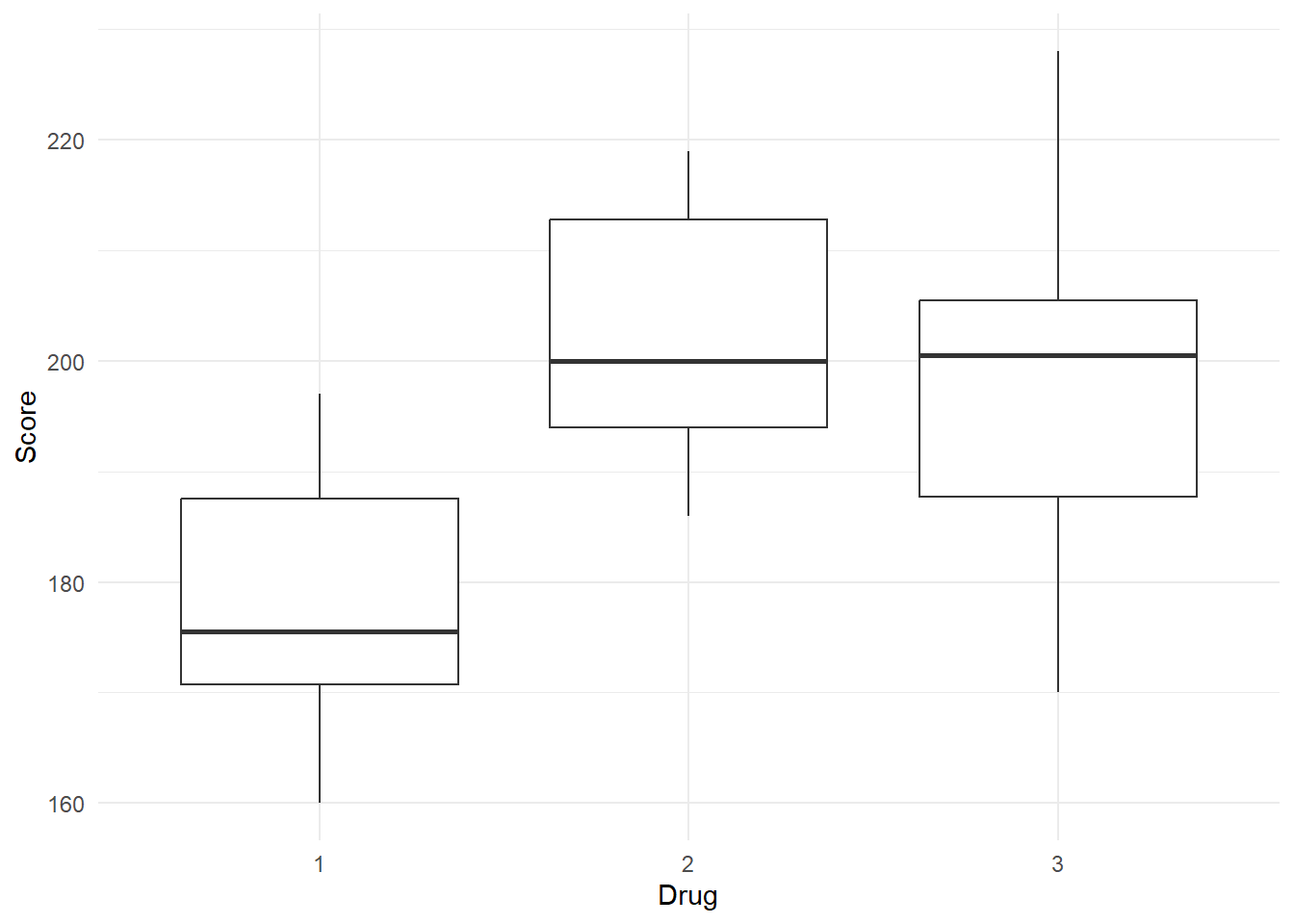
By Feedback
# Summary statistics
data %>%
group_by(Feedback) %>%
summarise(
n = n(),
min = min(Score),
max = max(Score),
median = median(Score),
mean = mean(Score),
sd = sd(Score)
) %>%
format_gt_tbl()| Feedback | n | min | max | median | mean | sd |
|---|---|---|---|---|---|---|
| 1 | 15 | 160 | 219 | 186 | 187 | 17.97 |
| 2 | 15 | 173 | 228 | 197 | 199 | 15.63 |
data %>%
ggplot(aes(x = Feedback, y = Score)) +
geom_boxplot() +
theme_minimal()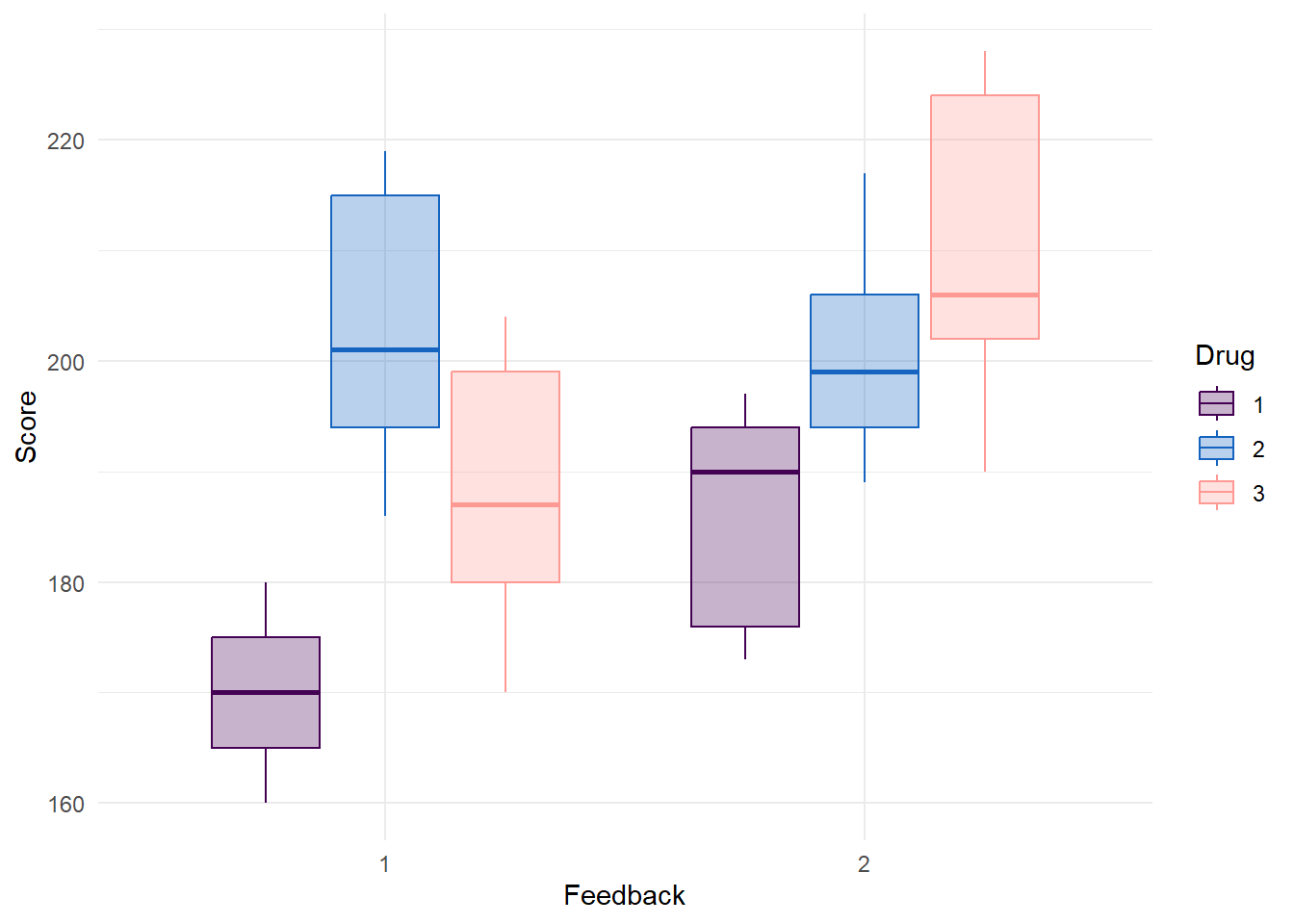
Plot Drug and Feedback
data %>%
ggplot(aes(x = Feedback, y = Score, color = Drug, fill = Drug)) +
geom_boxplot(alpha = 0.3) +
scale_color_manual(values = c("#440154FF","#1565c0", "#FF9993")) +
scale_fill_manual(values = c("#440154FF","#1565c0", "#FF9993")) +
theme_minimal()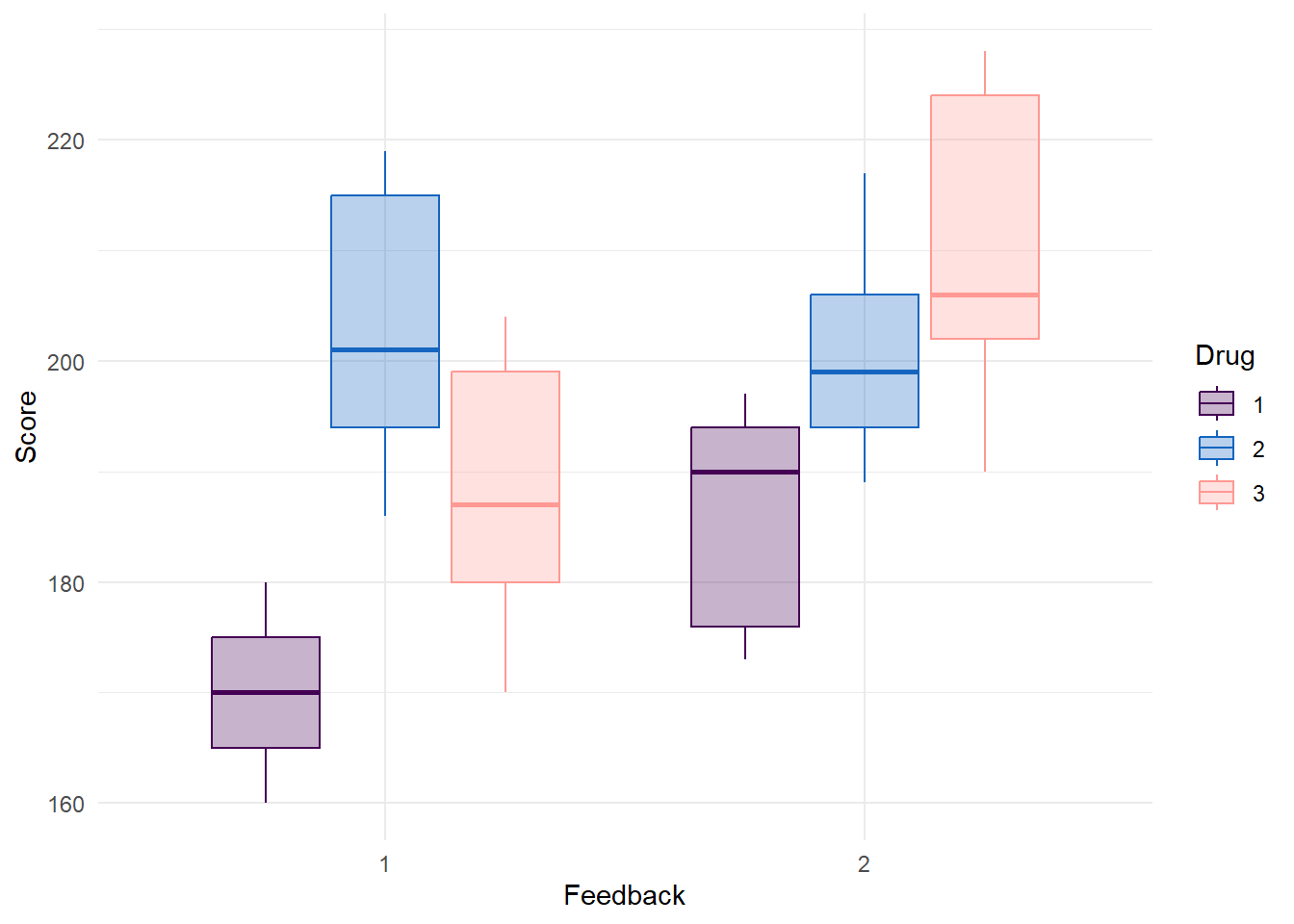
Summary statistics by Drug x Feedback (cell means)
# Summary statistics
summary_data <- data %>%
group_by(Feedback, Drug) %>%
summarise(
n = n(),
min = min(Score),
max = max(Score),
median = median(Score),
mean = mean(Score),
sd = sd(Score),
.groups = "drop"
)
summary_data %>%
format_gt_tbl()| Feedback | Drug | n | min | max | median | mean | sd |
|---|---|---|---|---|---|---|---|
| 1 | 1 | 5 | 160 | 180 | 170 | 170 | 7.91 |
| 1 | 2 | 5 | 186 | 219 | 201 | 203 | 13.91 |
| 1 | 3 | 5 | 170 | 204 | 187 | 188 | 13.84 |
| 2 | 1 | 5 | 173 | 197 | 190 | 186 | 10.84 |
| 2 | 2 | 5 | 189 | 217 | 199 | 201 | 10.93 |
| 2 | 3 | 5 | 190 | 228 | 206 | 210 | 15.81 |
Perform the two-way ANOVA
To perform a two-way ANOVA in base R, we can use the same lm() function used in the one-way ANOVA and input the formula as an argument.
# Specify an lm() model
model <- lm(Score ~ Feedback * Drug, data = data)
aov_tab <- car::Anova(model, type = "III")| Source | Sum Sq | Df | F value | Pr(>F) |
|---|---|---|---|---|
| (Intercept) | 144500 | 1 | 927.77 | < 0.001 |
| Feedback | 640 | 1 | 4.11 | 0.054 |
| Drug | 2730 | 2 | 8.76 | 0.001 |
| Feedback:Drug | 780 | 2 | 2.50 | 0.103 |
| Residuals | 3738 | 24 |
Effect size
eta_squared <- effectsize::eta_squared(car::Anova(model, type = 3), partial = TRUE)| Parameter | Eta2_partial | CI | CI_low | CI_high |
|---|---|---|---|---|
| Feedback | 0.146 | 0.95 | 0.000 | 1 |
| Drug | 0.422 | 0.95 | 0.148 | 1 |
| Feedback:Drug | 0.173 | 0.95 | 0.000 | 1 |
Interaction plot
ggplot(summary_data, aes(x = Feedback, y = mean, group = Drug, color = Drug)) +
geom_line(linewidth = 1) + # Connect means with lines
geom_point(size = 3) + # Add points for means
scale_color_manual(values = c("#440154FF","#1565c0", "#FF9993")) +
theme_minimal() +
labs(y = "Mean BP")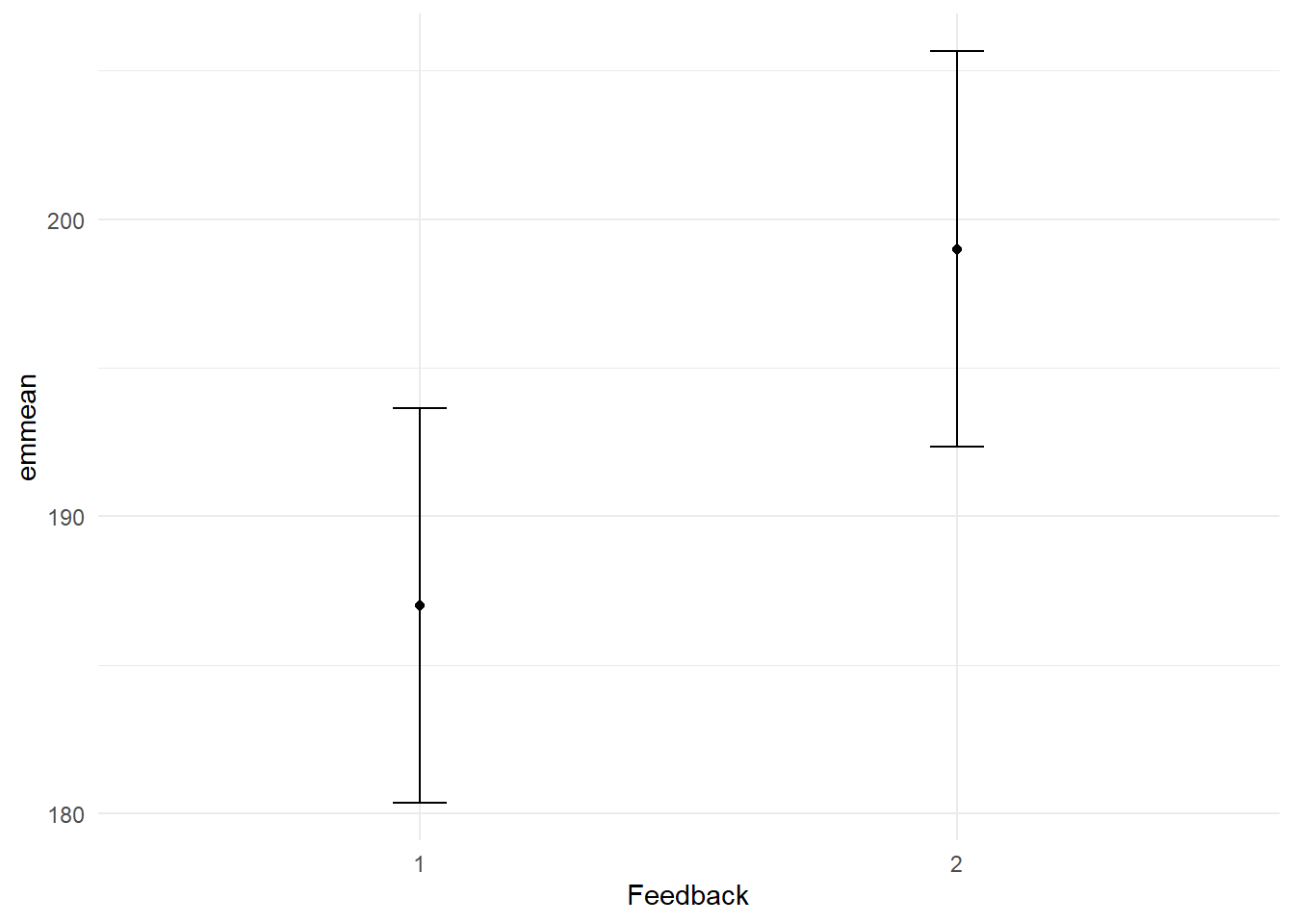
Tests of estimated marginal means
Suppose we wanted to know the overall difference between Feedback and no Feedback. Or we wanted to know the difference between between Drug 1 and Drug 2. Tests of estimated marginal means would be one way to accomplish this. In SAS these are referred to as least squares means (LS means).
Feedback
# Test of marginal means for Feedback
pwc <- emmeans::emmeans(model, ~ Feedback) %>%
pairs() %>%
as_tibble()
# Format the pwc table for display
pwc %>%
format_gt_tbl()| contrast | estimate | SE | df | t.ratio | p.value |
|---|---|---|---|---|---|
| Feedback1 - Feedback2 | -12 | 4.56 | 24 | −2.63 | 0.015 |
emms <- emmeans::emmeans(model, ~ Feedback) %>%
as_tibble()NOTE: Results may be misleading due to involvement in interactions# Print the estimated marginal means of Feedback
emms %>%
format_gt_tbl()| Feedback | emmean | SE | df | lower.CL | upper.CL |
|---|---|---|---|---|---|
| 1 | 187 | 3.22 | 24 | 180.35 | 193.65 |
| 2 | 199 | 3.22 | 24 | 192.35 | 205.65 |
# Plot the
emms %>%
ggplot(aes(x = Feedback, y = emmean)) +
geom_point() +
geom_errorbar(
aes(ymin = lower.CL, ymax = upper.CL),
width = 0.1) +
theme_minimal()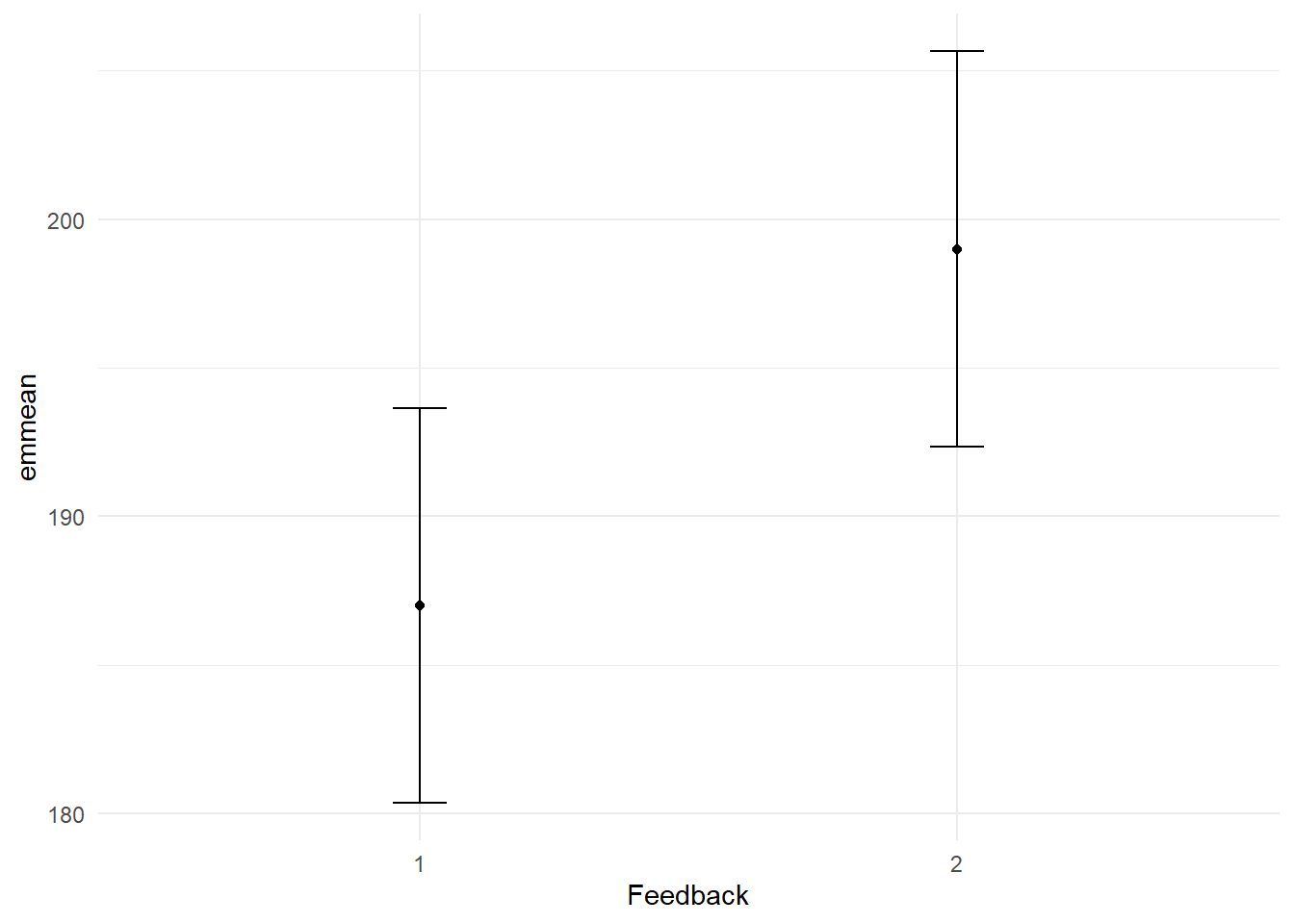
Drug
# Test of marginal means for Drug
pwc <- emmeans::emmeans(model, ~ Drug) %>%
pairs() %>%
as_tibble()
pwc %>%
format_gt_tbl()| contrast | estimate | SE | df | t.ratio | p.value |
|---|---|---|---|---|---|
| Drug1 - Drug2 | -24 | 5.58 | 24 | −4.30 | < 0.001 |
| Drug1 - Drug3 | -21 | 5.58 | 24 | −3.76 | 0.003 |
| Drug2 - Drug3 | 3 | 5.58 | 24 | 0.54 | 0.854 |
emms <- emmeans::emmeans(model, ~ Drug) %>%
as_tibble() NOTE: Results may be misleading due to involvement in interactions# Print the estimated marginal means of Drug
emms %>%
format_gt_tbl()| Drug | emmean | SE | df | lower.CL | upper.CL |
|---|---|---|---|---|---|
| 1 | 178 | 3.95 | 24 | 169.85 | 186.15 |
| 2 | 202 | 3.95 | 24 | 193.85 | 210.15 |
| 3 | 199 | 3.95 | 24 | 190.85 | 207.15 |
# Plot the means and the 95% CIs
emms %>%
ggplot(aes(x = Drug, y = emmean)) +
geom_point() +
geom_errorbar(
aes(ymin = lower.CL, ymax = upper.CL),
width = 0.1) +
theme_minimal()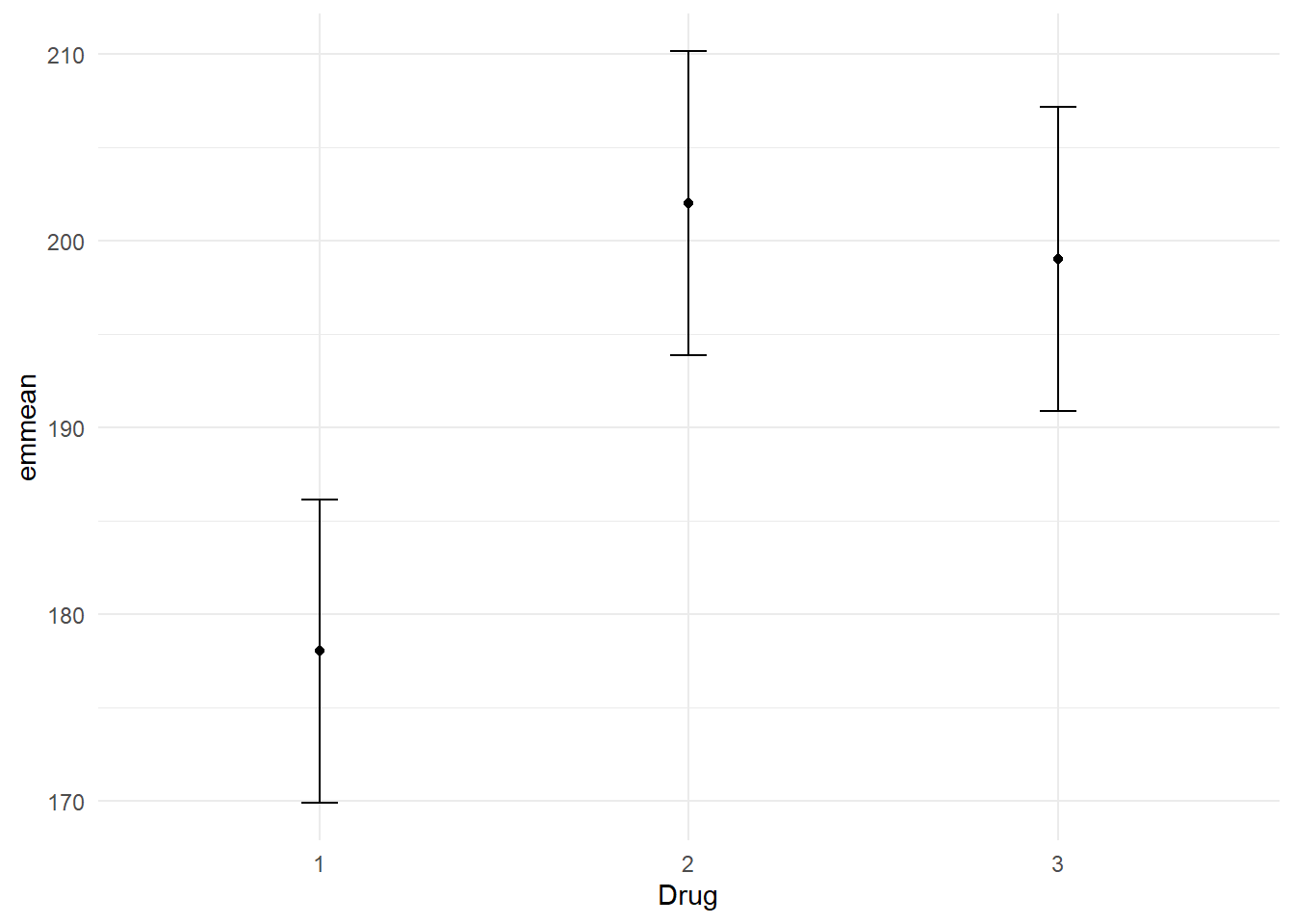
Interpretation
The omnibus Type III ANOVA revealed no significant interaction between Drug and Feedback, F(2, 24) = 2.50, p = .103, partial η² = .173. However, there were significant main effects. The main effect of Feedback was significant F(1, 24) = 6.93, p = .015, partial η² = .15. Post-hoc pairwise comparisons revealed an estimated reduction of 12 units in blood pressure with biofeedback, t = -2.63, p = 0.015, CI[3.062, 20.94]. Given the absence of a statistically significant interaction, we proceeded with tests of estimated marginal means to examine the main effects. The main effect of Drug was significant F(2, 24) = 10.98, p < .001, partial η² = .42. Post-hoc pairwise comparisons revealed an estimated reduction of 24 units in blood pressure for those on Drug 1 compared to Drug 2, t = -4.3, p < 0.001, CI[13.06, 34.94]. Similarly, those on Drug 1 had an estimated reduction of 21 units in blood pressure compared to those on Drug 3, t = -3.76, p = 0.003, CI[10.06, 31.94]. A comparison between Drug 2 and Drug 3 did not reveal a significant difference in blood pressure, t = 0.54, p = 0.854, CI[-13.94, 7.94].
If a significant interaction had been present, the appropriate next step would have been to test simple effects (e.g., Drug within levels of Feedback, or Fedback within levels of Drug). Nevertheless, the choice between marginal means and simple effects should ultimately be driven by the underlying research questions and theoretical considerations of the research question of interest.
References
Maxwell, Scott, Harold Delaney, and Ken Kelley. 2020. AMCP: A Model Comparison Perspective. https://CRAN.R-project.org/package=AMCP.
Maxwell, Scott E, Harold D Delaney, and Ken Kelley. 2017. Designing Experiments and Analyzing Data: A Model Comparison Perspective. Routledge.
Wickham, Hadley. 2019. Tidyverse: Easily Install and Load the ’Tidyverse’. https://CRAN.R-project.org/package=tidyverse.